Research
Research in my laboratory is focused on exploring the mechanisms that allow Staphylococcus aureus to cause disease in humans. Infections caused by S. aureus, and in particular those caused by the highly drug resistant form, methicillin resistant S. aureus (MRSA), are a growing problem in the United States and very few antibiotics remain effective in treating diseases caused by this bacteria. Understanding the pathogenesis of S. aureus is critical to aid the development of effective vaccines and treatments.
RNA-based regulation in Staphylococcus aureus
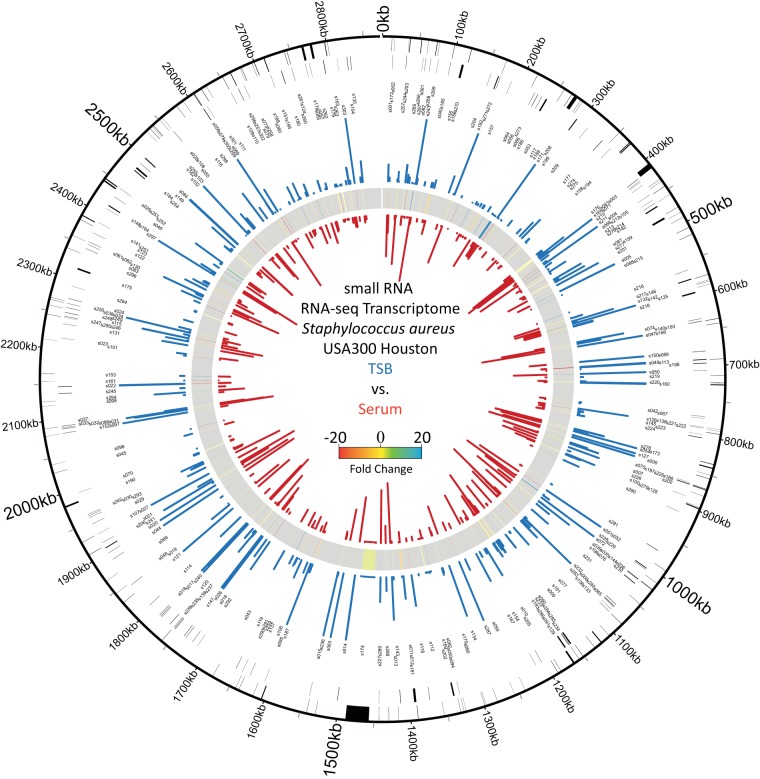
Regulatory RNAs (also called small RNAs or sRNAs) greatly outnumber regulatory proteins in the S. aureus cell, yet they remain poorly understood and severely understudied. One factor that has hampered the study of sRNAs in S. aureus has been the lack of a clear nomenclature system and their absence from genome annotation files. To facilitate improved study of sRNAs in S. aureus we performed a comprehensive re-annotation of the S. aureus genome to identify and annotate all known sRNAs. We can now perform RNAseq experiments and, using the updated genome file as a reference, search for previously undiscovered sRNA genes, and monitor global sRNA gene expression. This has opened the door to multiple lines of investigation in my lab centered around the role of RNA molecules in gene regulation. We have identified a trans acting sRNA (Teg41) that controls production of the cytolytic αPSM peptide(s) and also investigated cis acting thermoswitches that control translation of S. aureus genes in response to temperature.
Temperature-based gene regulation in Staphylococcus aureus
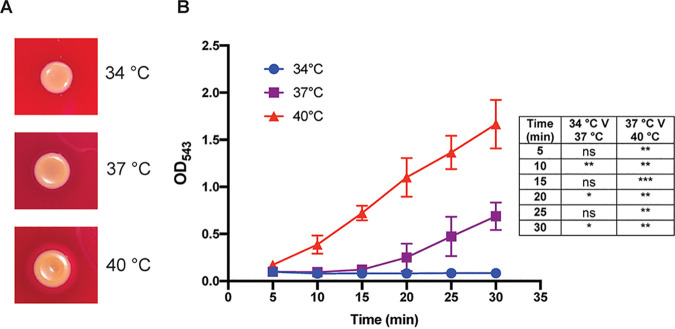
Many pathogenic bacteria use temperature as a signal to regulate expression of virulence genes once they have entered the human host. During its transition from commensal to pathogen S. aureus undergoes a small but significant change in temperature from 34˚C to 37˚C. We have investigated how this change in temperature alters the transcriptional and proteomic landscape of the bacteria and shown that S. aureus is subject to a large degree of post-transcriptional temperature dependent gene regulation. RNA molecules appear to be at the heart of this regulation with sRNAs, thermoswitches and RNA thermometers all playing a role.
Staphylococcus aureus Extracellular Vesicles (EVs)
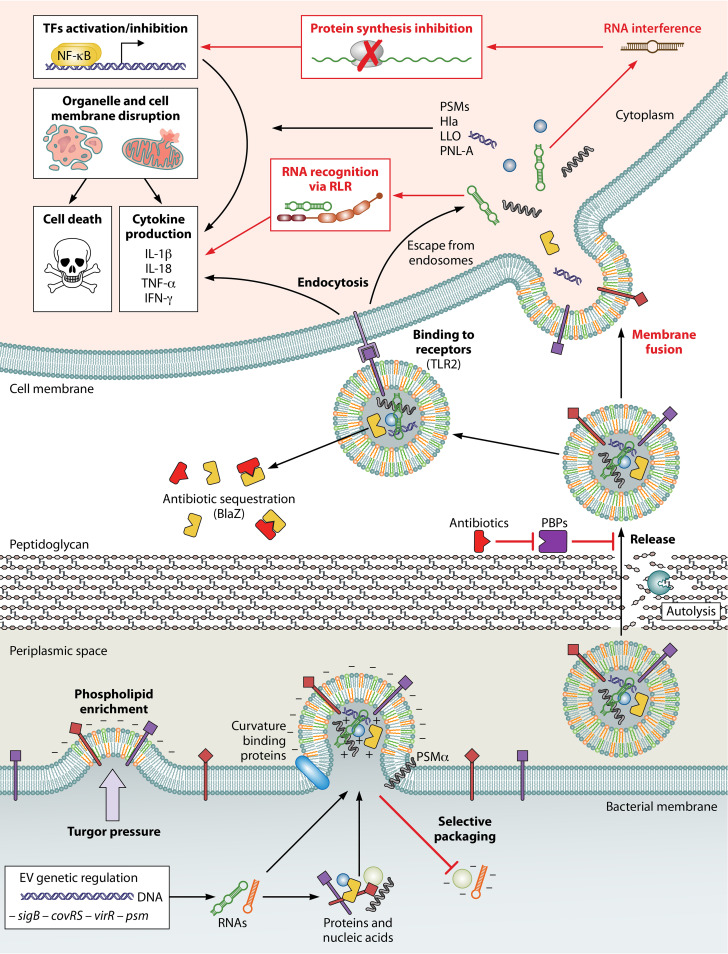
S. aureus, like most other bacterial species, produces and secretes extracellular vesicles (EVs). EVs are nano sized spherical lipid bilayers containing a variety of biomolecules (including proteins, lipids, DNA and RNA). Work in our lab has shown that the composition and number of EVs produced by S. aureus depends on temperature. The cytotoxicity of EVs towards human cells also appears to vary at different temperatures and EVs produced at different temperatures have different effects on different cell types. We are investigating the molecules and mechanisms responsible for these phenotypes. Interestingly the RNA content of EVs appears to increase at lower temperatures raising the possibility that some of the EV related phenotypes could be RNA dependent.
Peptidyl-prolyl cis-trans isomerases (PPIases) and virulence
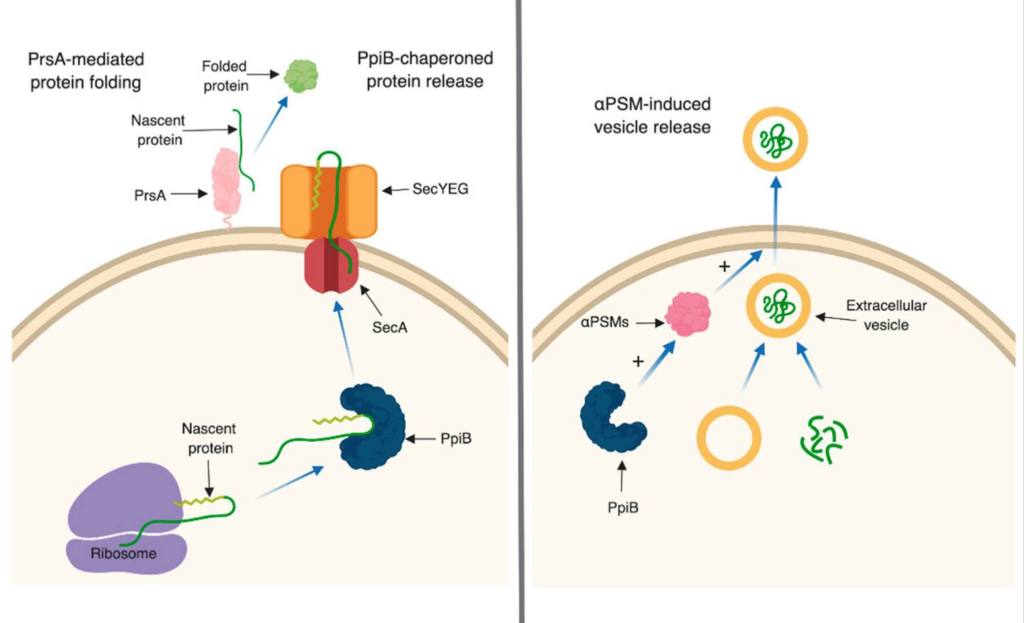
PPIase enzymes are best studied in Gram-positive bacteria for their role in folding proteins after they are secreted from the cell, however they can also have additional roles and functions. Our research focuses on two S. aureus PPIases, PrsA and PpiB. PrsA is a cell-membrane anchored lipoprotein that appears to influence S. aureus pathogenesis by folding secreted proteins. PpiB also influences secretion and activity of S. aureus virulence factors, however it does so from inside the bacterial cell. Recent results suggests that PpiB may be influencing secretion in S. aureus either through interactions with Sec system components or by playing a role in the release of extracellular vesicles.